Conditional Diffusion
Mask diffusion decomposes the generation of the segmentation mask into a step-by-step denoising process:
- forward
- backward
use
to estimate
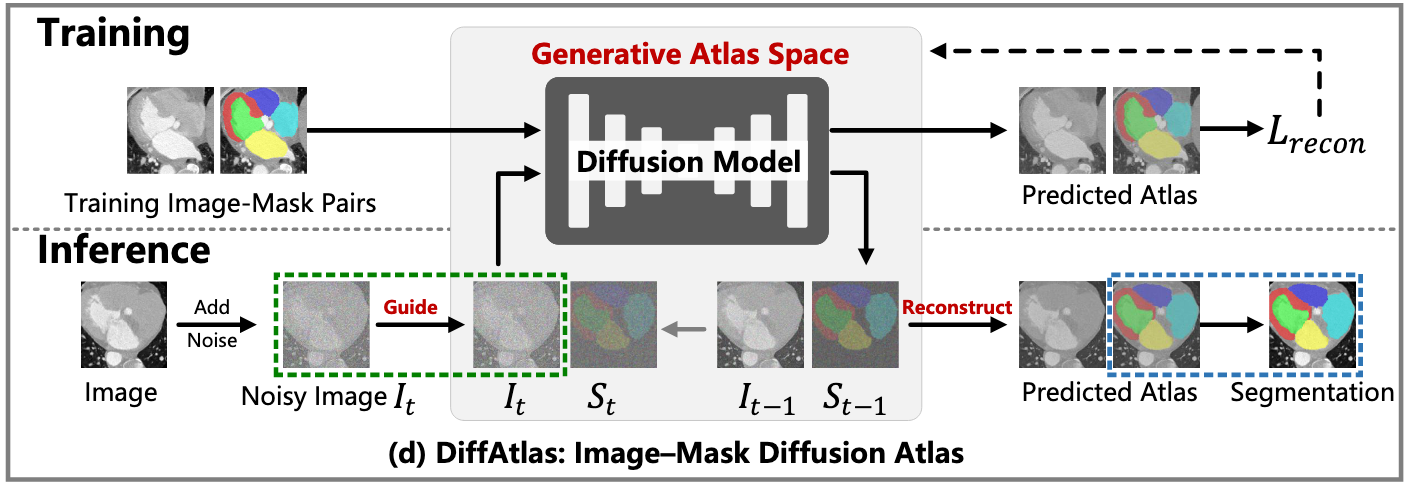
DiffAtlas: training
DiffAtlas models the input image and segmentation mask as a pair
Details
# self.denoise_fn is a UNet3D
recon = self.denoise_fn(
**dict(
x=torch.cat((x_noisy, m_noisy), dim=1),
time=t,
cond=cond,
**kwargs
)
)
x_recon = recon[:,0,:,:,:]
m_recon = recon[:,1:(recon.size()[1]),:,:,:]
if self.loss_type == 'l1':
loss = F.l1_loss(noise_x, x_recon) + F.l1_loss(noise_m, m_recon)
elif self.loss_type == 'l2':
loss = F.mse_loss(noise_x, x_recon) + F.mse_loss(noise_m, m_recon)
DiffAtlas represents the mask S using a signed distance function (SDF) to improve the capture of fine anatomical details and ensuring smooth transitions between regions, which is based on FlowSDF.
DiffAtlas: inference

- Randomly initialized noisy image-mask pair
- Refines it iteratively through the reverse diffusion process over T timesteps.
- At each timestep
- At each timestep
Visualization
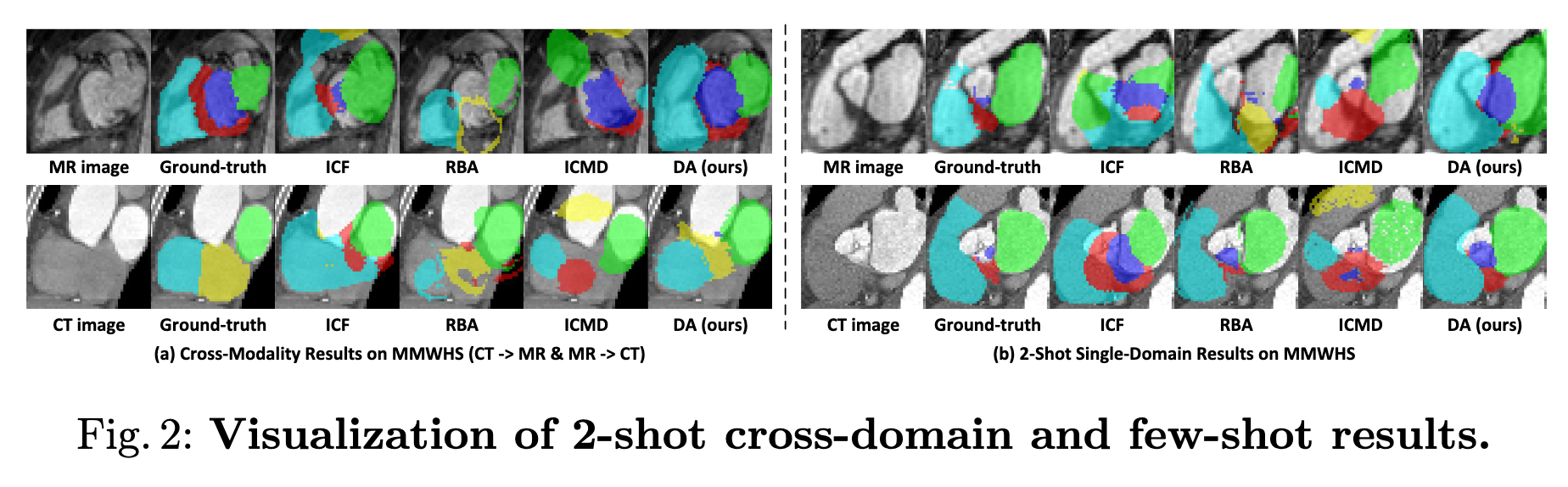
Method Comparison:
- ICF (ImageConditional Feedforward): nnU-Net
- RBA (Registration-Based Atlas): CMMAS
- ICMD (Mask Diffusion): MedSegDiffv2
- DA (Ours): DiffAtlas
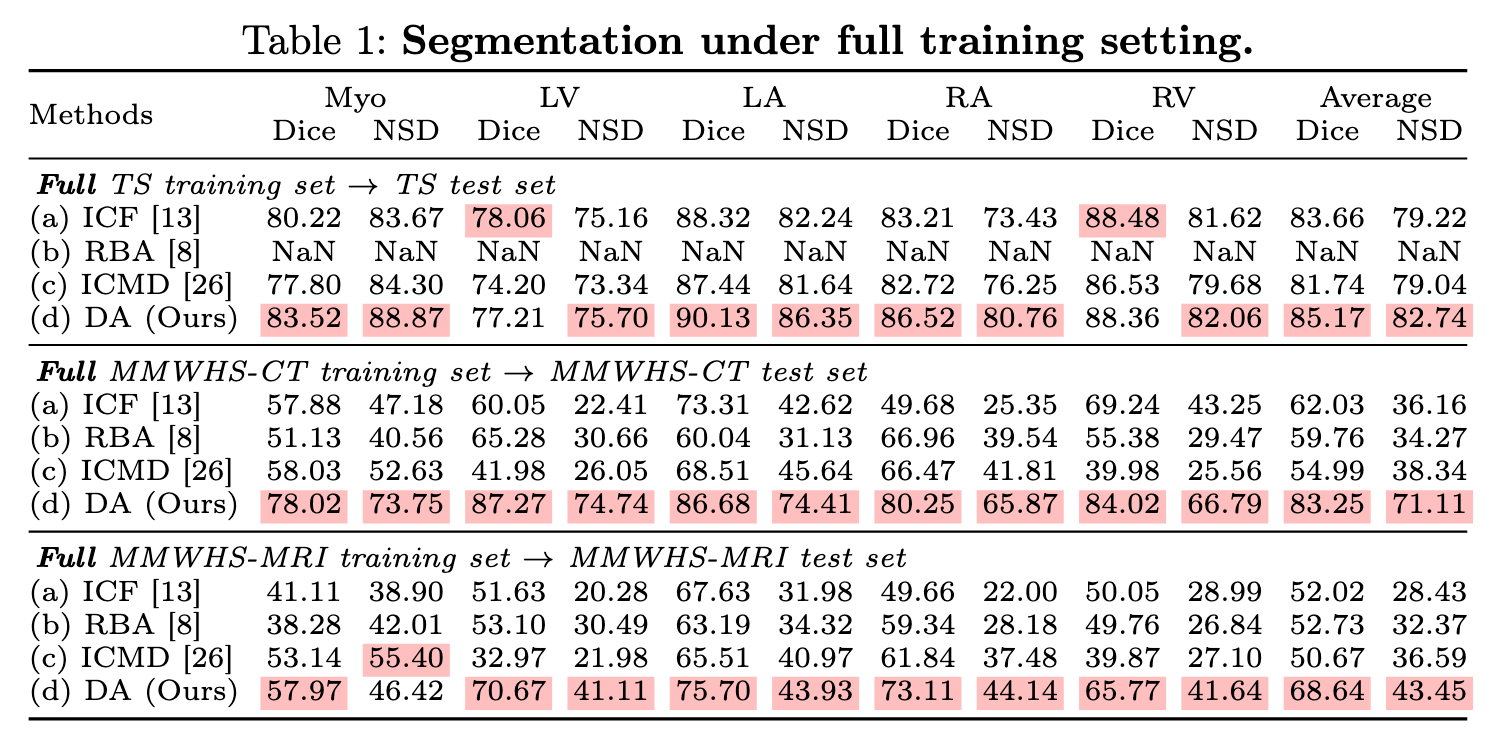
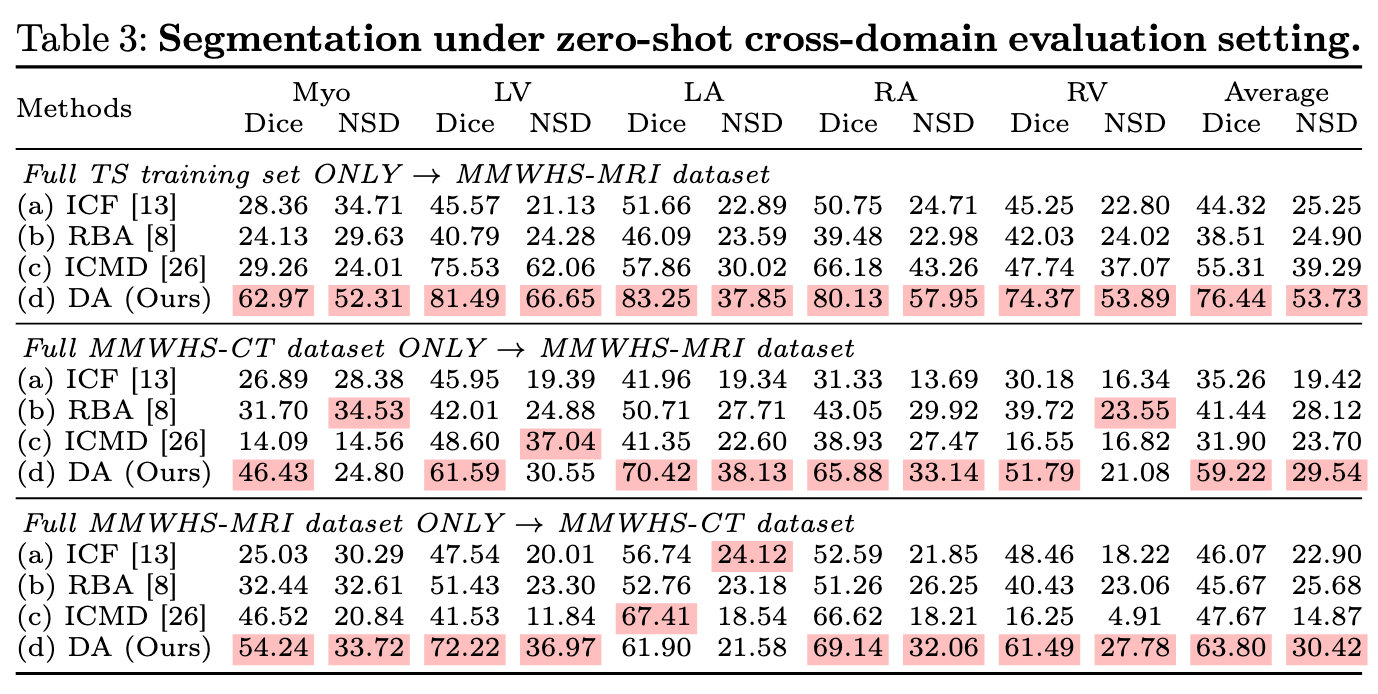
Experiments 1: full training setting
With TotalSegmentator(746 CT) and MM-WHS(20CT + 20MR) dataset, train:test = 8:2.
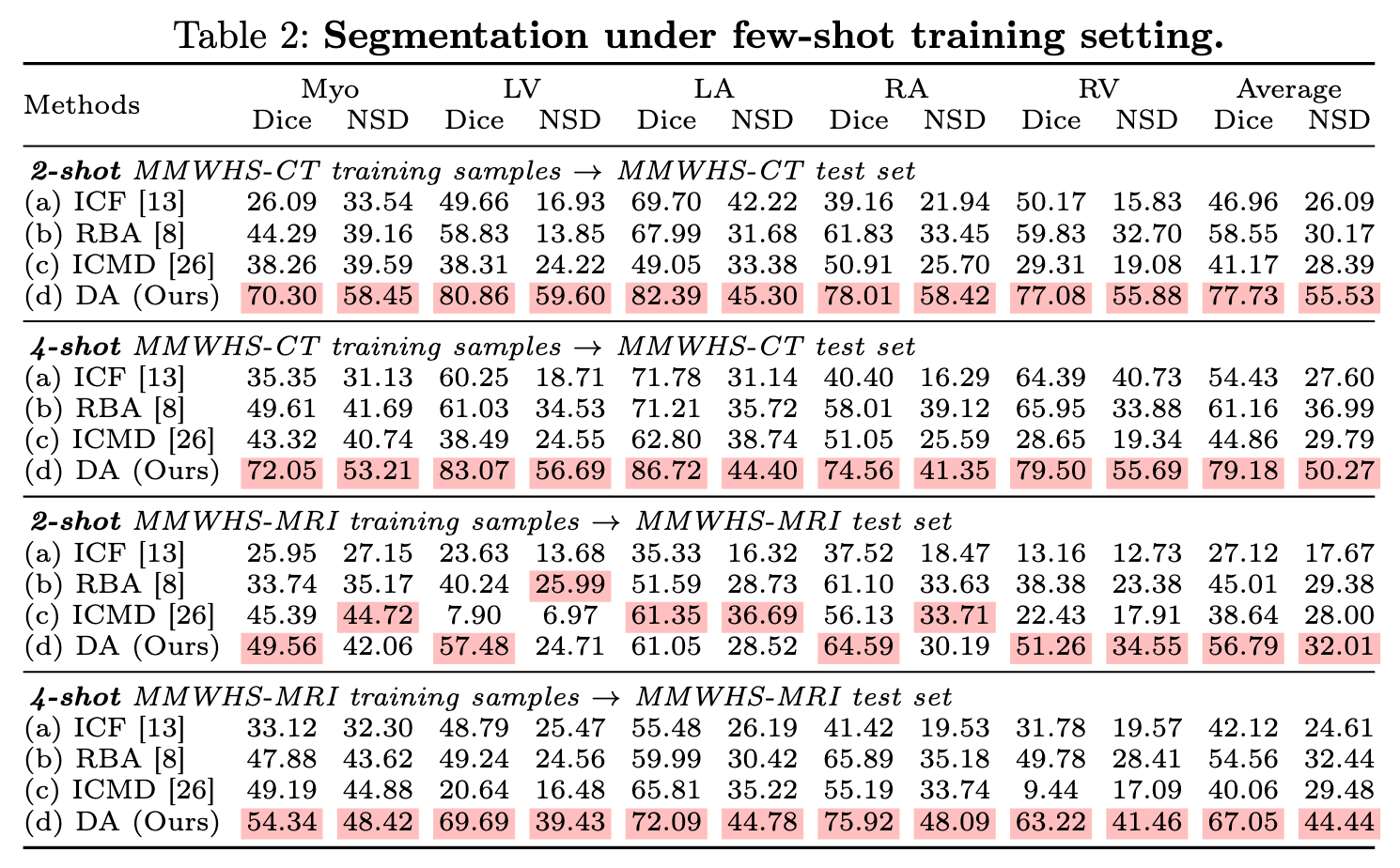
Experiments 2: few shot
- 2-shot (2 training samples) and 4-shot (4 training samples).
- Test samples remain the same.
Experiments 3: cross modality
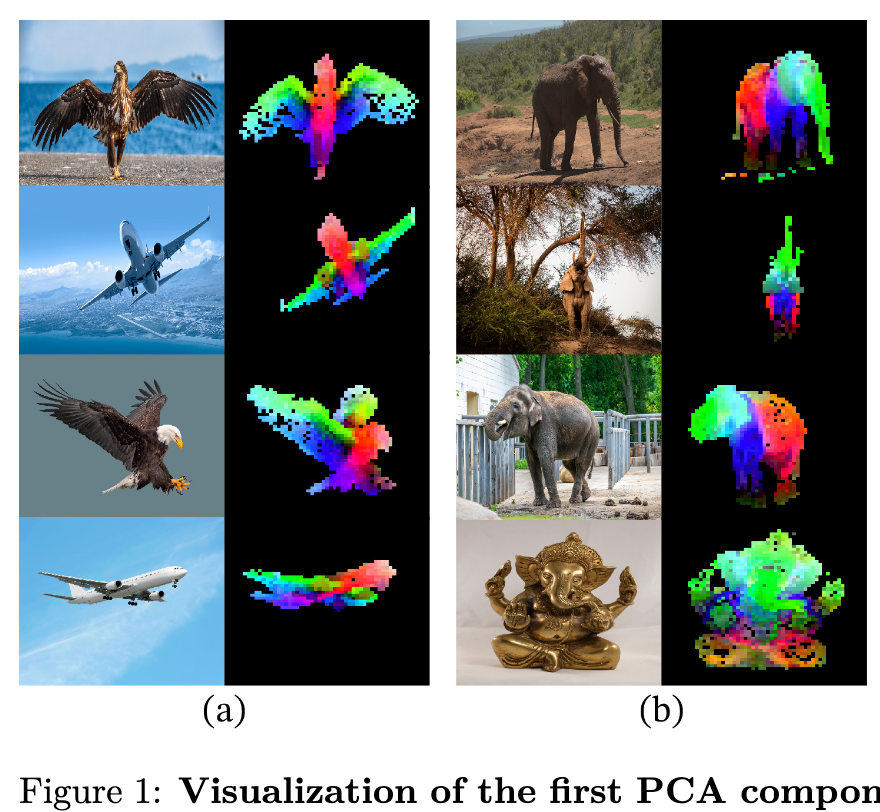
Introduction

There are two line of works to tackle unsupervised anomaly detection (UAD) problems.
- reconstruction-based approaches, where a generative model is trained to reconstruct normal images.
- model the distribution of the features extracted from normal samples.
DINO v2
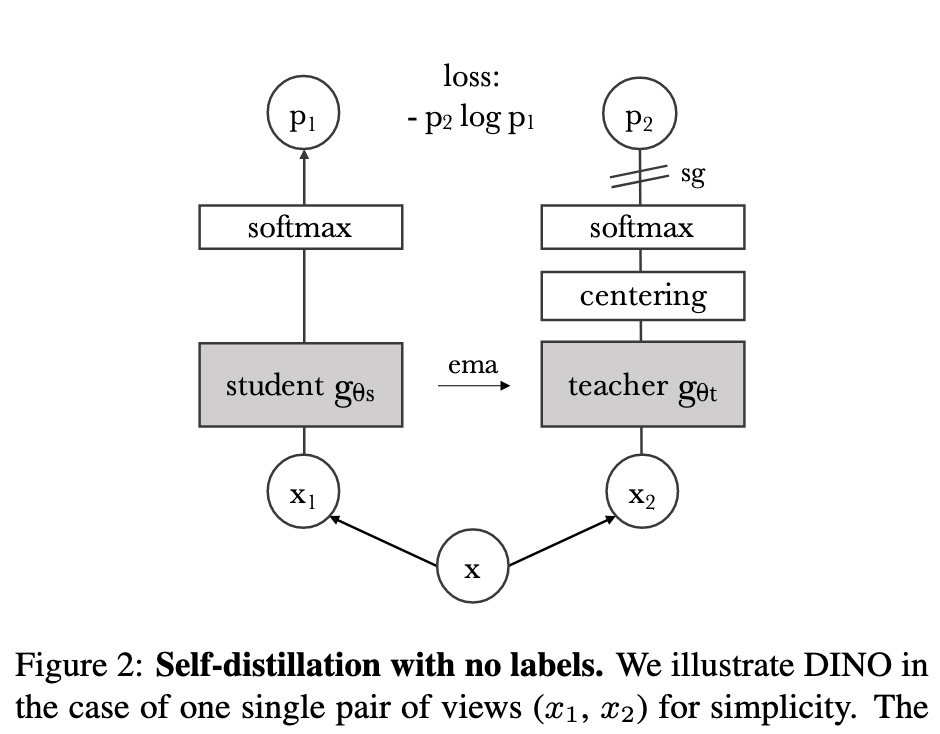
- Left: DINO framework
- Right: DINOv2, more powerful and robust for many tasks.
AnomalyDINO

AnomalyDINO-DPMM
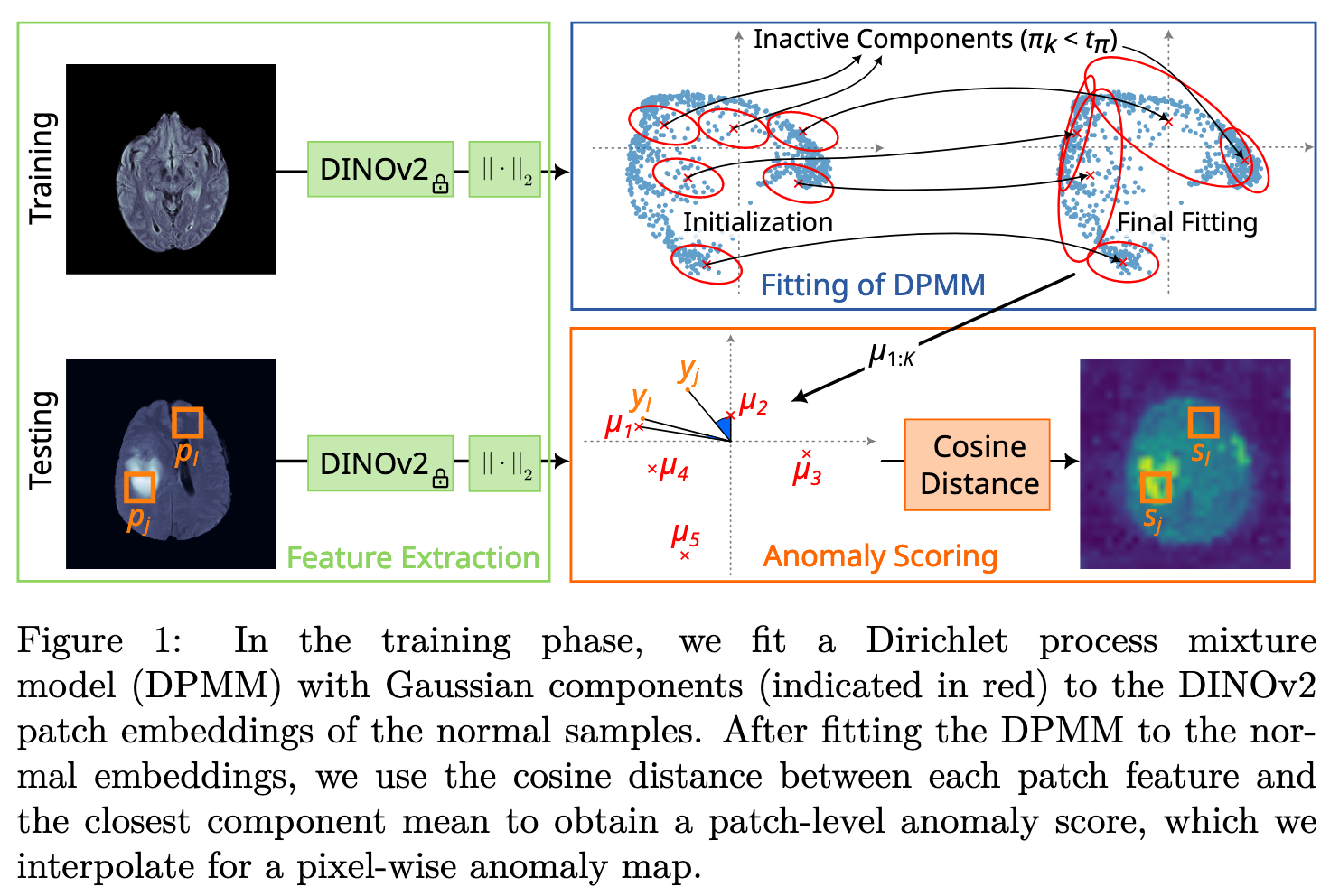
Intuition
My personal insight
What AnomalyDINO-DPMM have done is similar to vector database indexing (i.e. some kind of compression or clustering, aimed to acculerate vector comparison):
DPMM
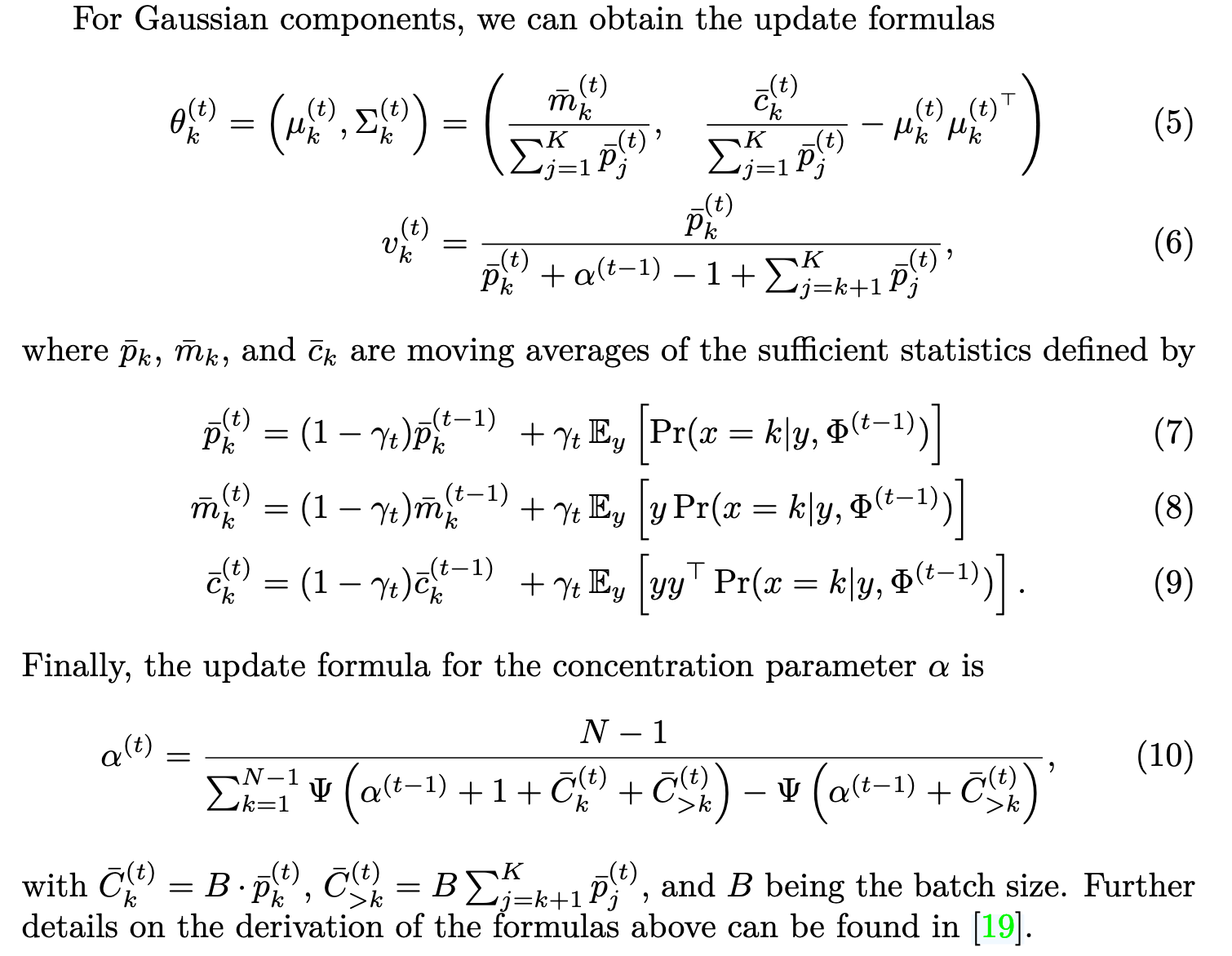
DPMM is a Gaussian mixture model without a fixed number of components that instead determines the necessary number of components based on the data, allowing for a more flexible, data-driven approach.

EM iteration for DPMM
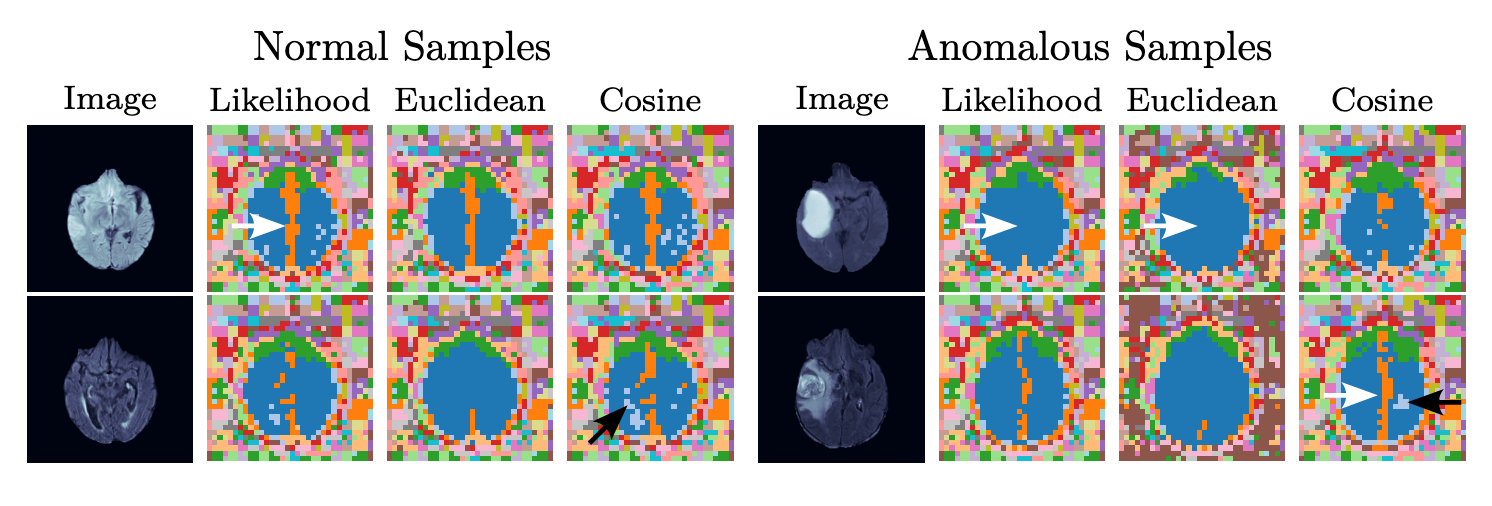
Anomaly Scoring
-
likelihood:
-
Euclidean distance:
-
Cosine similarity:
To suppress all components with vanishing weights, we only consider these k:
Experiments

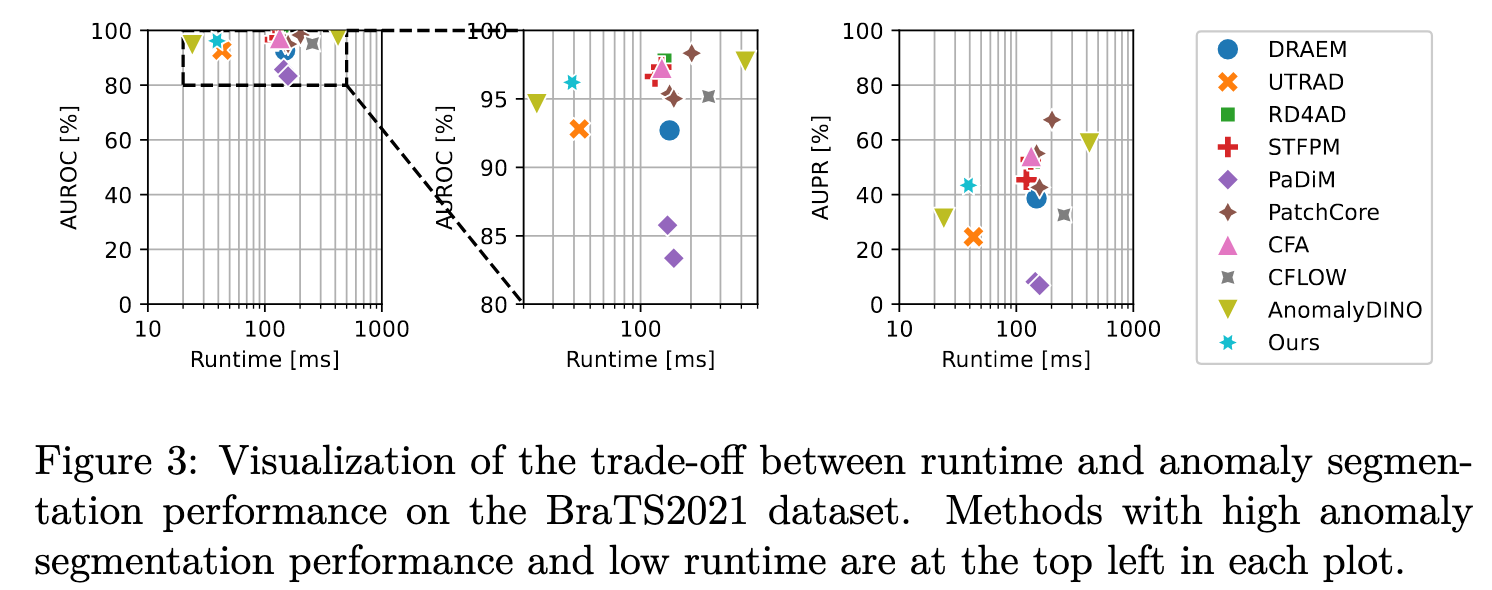
Efficiency-Performance Trade-off

THANKS